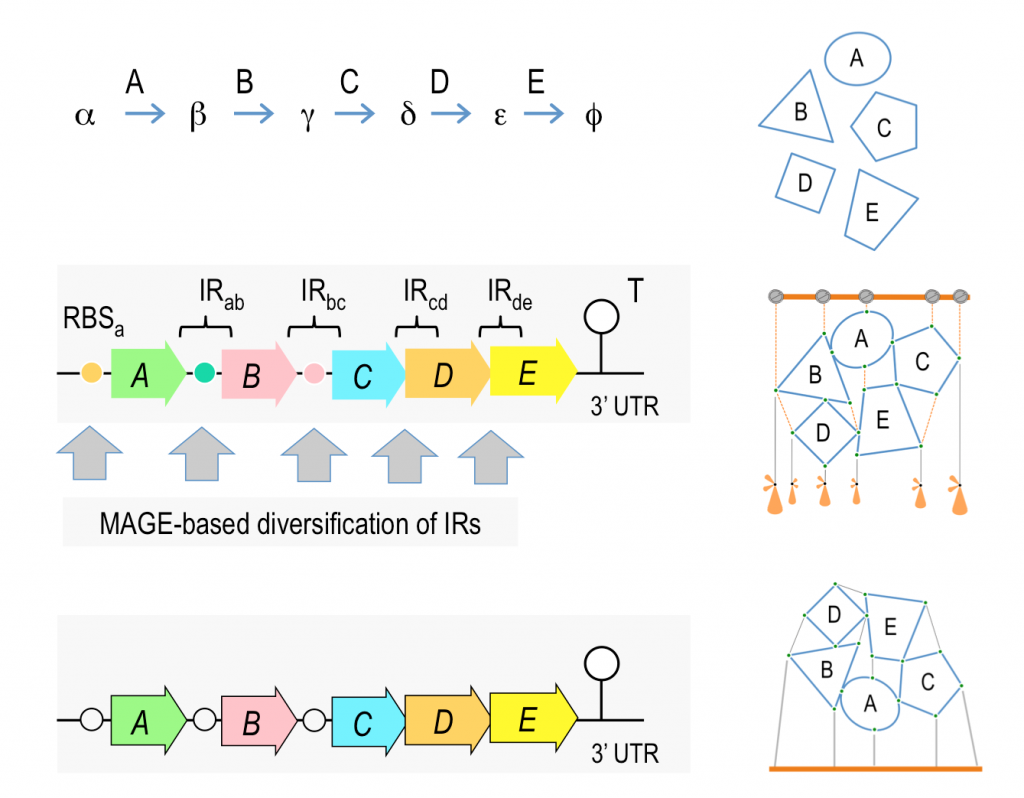
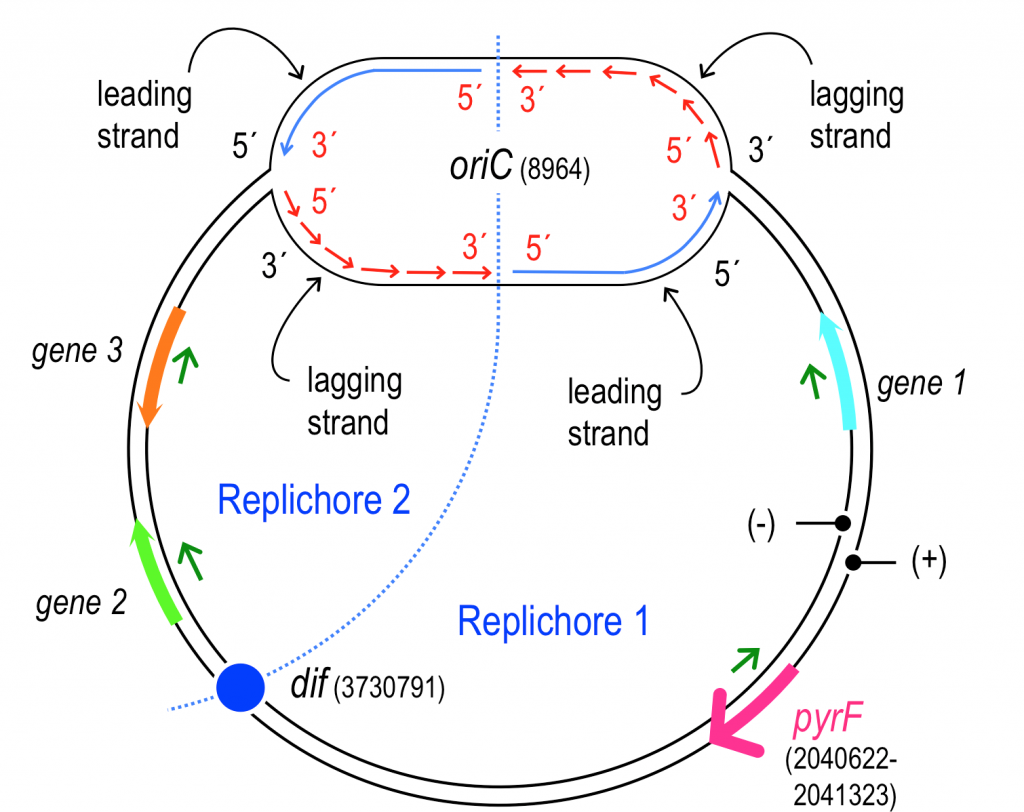
Why Pseudomonas putida?
The workhorse and experimental system par excellence in our Laboratory is the Gram-negative soil bacterium and plant-colonizer Pseudomonas putida, in particular the strain called KT2440. But in fact, we are not as interested in the lifestyle of this bacterium in its natural niches as on its awesome physiological strength and stress-tolerance properties. We try to domesticate and repurpose these features with a biotechnological aim with the tools of contemporary Synthetic Biology. While Escherichia coli constitutes an optimal model in Biology, many reactions and processes of environmental and industrial interest proceed with the accumulation of toxic intermediates that E. coli can hardly cope with. In contrast, P. putida seems to fulfill nearly all conditions required in an ideal cell factory, as this bacterium adapt easily to very different physicochemical and nutritional niches and it is well equipped to endure both endogenous and exogenous stresses. The origin of such heftiness is to be found in its central biochemical network, which is well geared to produce the metabolic currency required to overcome all types of physical and chemical insults. Our starting point is thus a natural bacterium that we have tried over the years to tame for our own sake and to redesign in many different ways.