The CRISPR page at CNB
by Lluís Montoliu (CNB-CSIC)
with help and comments from Davide Seruggia (Boston Children's Hospital) and Francis J.M. Mojica (Universidad de Alicante)
LAST UPDATED: 25 July 2019
Table of Contents for this Web page
Which are the basic papers describing the use of CRISPR-Cas9 technology?
Additional papers worth reading illustrating the many uses of CRISPR-Cas9 technologies
Brief history of CRISPR: Who discovered this system in bacteria?
Who is Who in the CRISPR-Cas World
CRISPR publications from Francis Mojica
Videos from CRISPR talks and seminars delivered by Francis Mojica
CRISPR publications from Lluís Montoliu
Videos from CRISPR talks and seminars delivered by Lluís Montoliu
NHEJ inhibitors: boosting the HDR pathway
Where can I get full protocols for using CRISPR-Cas-related technologies?
Successful Knock-In strategies with CRISPR-Cas tools
CRISPR experiments in livestock
CRISPR and gene drive approaches
Anti-CRISPR and Cas inhibitors
Commercial resources for CRISPR-Cas-related reagents
Where can I discuss with other colleagues about the use of CRISPR-Cas9 techniques?
Global sites of CRISPR information, publications & commentaries
Beyond CRISPR-Cas: Ago (Argonaute) No evidences of gene-editing activity
.
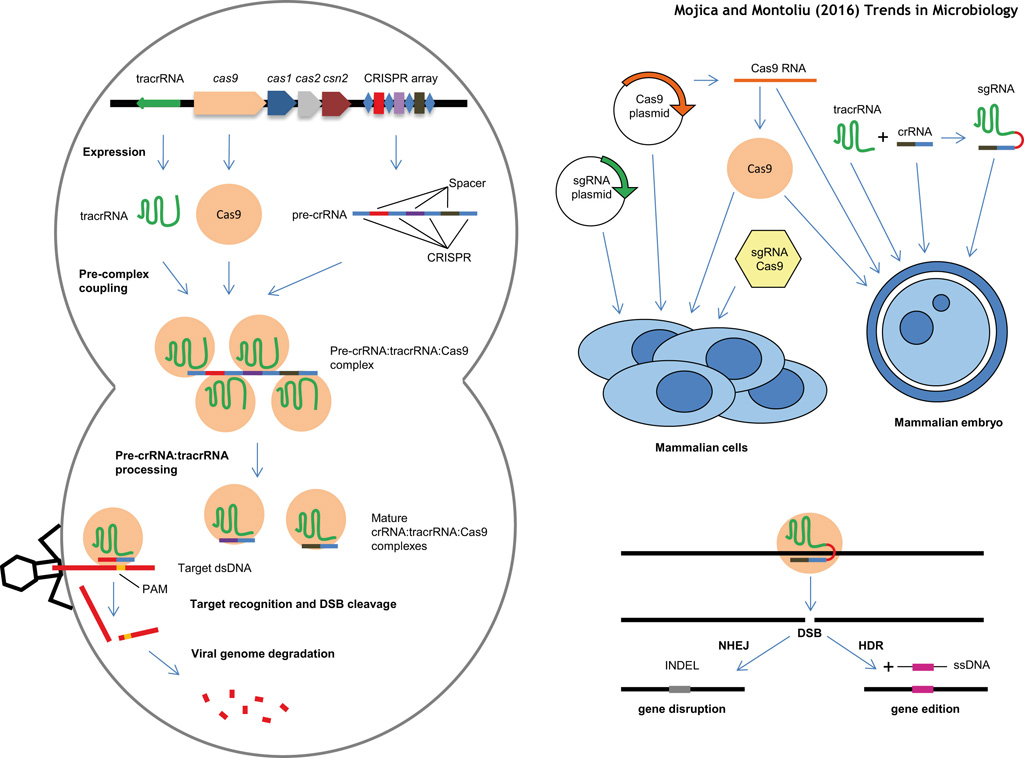
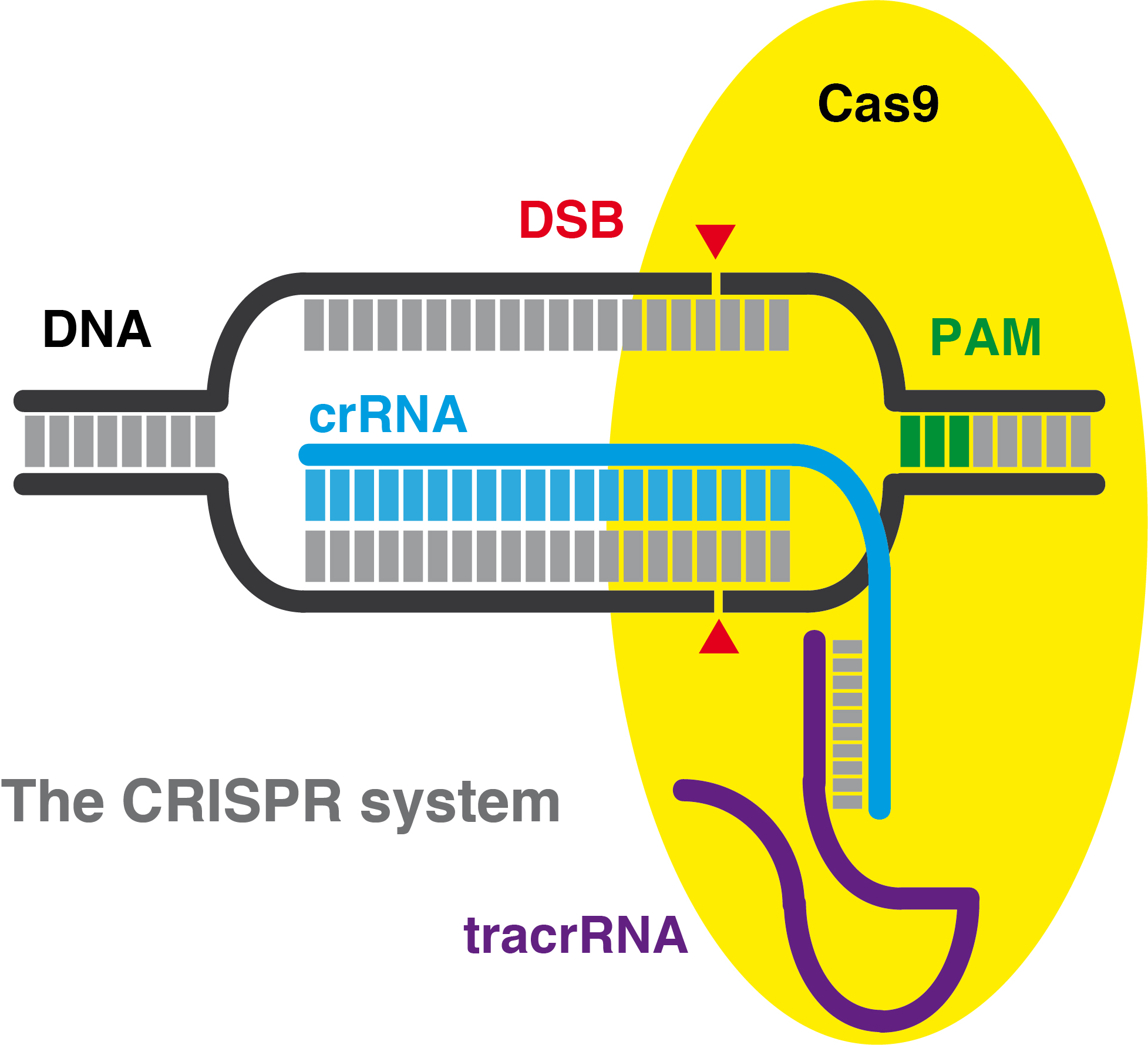
| CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) sequences and Cas (CRISPR-associated) proteins are the two elements of an ancient defence prokaryotic adaptive restriction system conserved in bacterial genomes. While CRISPRs represent the memory of the system, a repository of short, directly repeating nucleotide sequences flanked by short unique unique DNA fragments, acquired from previous infections, Cas proteins are the actual effectors, that are able to process the CRISPR sequences into small RNAs, and to cleave those infectious DNA molecules that perfectly match those CRISPR-derived RNAs. To translate a complex prokaryotic system into a simple genome editing tool, the crRNA (CRISPR RNA) and the tracrRNA (trans-activating crRNA) have been fused in a synthetic small guide RNA (sgRNA), composed by a hairpin RNA structure, resembling the tracrRNA, linked to a 20 bp sequence homologous to the target DNA. Out of all the CRISPR-associated proteins, Cas9 is the final effector, capable of complexing and cleaving both strands of a DNA molecule upon detecting a typical Watson&Crick homologous base pair match with the sgRNA. Therefore the CRISPR-Cas system is often referred as CRISPR/Cas9. Genome engineering by these RNA-programmable Cas9 nucleases have broad applications in biology, biomedicine and biotechnology. The microbiologist Emmanuelle Charpentier and the structural biologist Jennifer Doudna, along with the rest of their colleagues co-authoring the first seminal publications on this subject, are the ones to be credited for having investigated and brought this amazing prokaryotic tool to the attention and for the benefit of the eukaryotic world. |
|
CRISPR-Cas technology and applications (video by Le Cong from Feng Zhang's lab at MIT/BROAD institute) This video has been made possible thanks to Addgene and is part of this series . . . Scheme illustrating the CRISPR-Cas9 mechanism of action (by Lluís Montoliu ©) .
. Scheme illustrating the two repair pathways occurring after CRISPR-Cas9 action (by Lluís Montoliu ©) . |
Which are the basic papers
describing the use of CRISPR-Cas9 technology?.![]()
There are already many papers, reviews and
reports describing the CRISPR-Cas9 technology. These few papers
below are our recommended "starting set" of "MUST-READ" papers,
listed as they were published, worth reading before starting using
this technology. Of course, additional publications appear
constantly and, hence, it is advisable to regularly review the most
recent literature on this subject.
|
Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR–Cas9-mediated mutagenesis
Seruggia et al. Nucleic Acids Research (2015)
Additional papers worth
reading illustrating the many uses of CRISPR-Cas9 technologies.![]()
These are additional papers we have found
useful and interesting to read to illustrate the many uses of the
new CRISPR-Cas9 technology for genome edition purposes. This list is
non-exhaustive, nor it represents necessarily the best papers
published. It is simply a list of recent publications which have
been selected according to the novelty of the technical or
scientific approaches reported.
|
Francis Mojica (University of Alicante) presenting "the origins of the CRISPR-Cas systems"
in a seminar at CNB-CSIC, Madrid, on 14 October 2015, picture by Lluis Montoliu
.
Francis Mojica (University of Alicante) -right- with Lluis Montoliu
Madrid, 14 October 2015, picture by Lluis Montoliu
.
.Brief
history of CRISPR: Who discovered this system in bacteria?.![]()
There are the first published publications
and books
describing CRISPR in bacteria and archeas, eventually leading to the current
scenario where CRISPR are applied universally.
|

Lluis Montoliu and Francis Mojica at BioSpain-2016 in Bilbao
picture by Lluis Montoliu
.
Who is Who in the CRISPR-Cas
World.![]()
|
This is a non-exhaustive collection of pictures with some of the many researchers that have contributed significantly with their studies to our current understanding of the CRISPR-Cas systems in prokaryotes and their applicaton for genome editing purposes in eukaryotes. . |
|
Francis Mojica
|
Rodolphe Barrangou North Carolina State Univ, Raleigh, USA
|
Philippe Horvath DuPont Nutrition and Health, France
|
Luciano Marraffini The Rockefeller Univ, New York, USA
|
|
John van der Oost Wageningen University, The Netherlands
|
Emmanuelle Charpentier MPI for Infect. Biol., Berlin, Germany
|
Jennifer Doudna Univ California Berkeley, CA, USA
|
Virginijus Siksnys
|
|
Feng Zhang
|
George Church Harvard Med School, Boston, MA, USA
|
Rudolf Jaenisch Whitehead Inst, Cambridge, MA, USA
|
J. Keith Joung Mass Gen Hosp, Charlestown, MA, USA
|
.
. |
Francisco Juan Martínez Mojica (Francis Mojica), Universidad de Alicante. Fotografía: Roberto Ruiz. Taller de Imagen (UA)
CRISPR
publications from Francis Mojica ![]()
These are CRISPR publications from Francis
Mojica and/or his lab, published in diverse scientific journals. PDF copies from all these papers are
available upon request.
Francis Mojica surrounded by UAM Master students after delivering a class on CRISPR systems (course 2017-18) . |
Francis Mojica impartiendo una conferencia en los Encuentros con la Ciencia-Málaga
Videos
from CRISPR talks and seminars delivered by Francis Mojica
![]()
These are some of the videos from CRISPR
talks, seminars and interviews delivered by Francisco Juan Martínez Mojica,
Francis Mojica
(Universidad de Alicante) on his discovery of the CRISPR systems in
prokaryotes. Some of these videos are in
Spanish.
|
Lluís Montoliu, fotografía: Roberto Pérez-El Mundo
CRISPR
publications from Lluís Montoliu ![]()
These are CRISPR publications from Lluís
Montoliu and/or his lab, published in diverse scientific journals,
webs, blogs or popular science articles, as they appeared in
newspapers or diverse journals. PDF copies from all these papers are
available upon request. Some of these popular science articles are
in Spanish.
|

Lluís Montoliu, conferencia en El Foro de Madrid
Videos
from CRISPR talks and seminars delivered by Lluís Montoliu
![]()
These are some of the videos from CRISPR
talks, seminars and interviews delivered by Lluís Montoliu on the
subject of gene editing mediated by CRISPR. Some of these videos are
in Spanish.
|
NHEJ inhibitors
and other strategies for boosting the
HDR pathway.![]()
There are the first published publications
reporting the use of a variety of NHEJ inhibitors in order to boost
the HDR pathway when using a CRISPR-Cas9 approach, in order to
improve the efficiency and success of knock-in/genome edition/substitution
features versus the standard, default and far more effective NHEJ
repair system, usually leading to the accumulation of indels.
|
Alternative methods to embryo
microinjection to generate genome-edited mice with CRISPR-Cas9.![]()
These articles reported
the use of embryo electroporation as a valid alternative method to
pronuclear/citoplasmic microinjection of CRISPR-Cas9 reagents. The
main difference is whether embryos are first obtained, their zona
pellucida digested before electroporation and then subsequently
transferred into recipient pseudopregnant females, as described by
Qin et al. 2015; Hashimoto&Takemoto 2015; Qin et al. 2016, or whether the
electroporation step is done in situ, in oviduct, as in the GONAD
method (Takahashi et al. 2015), thus avoiding both the
microinjection and the embryo-transfer steps. A new article has been
added combining ultrasuperovulation with IASe reagent,
cryopreservation methods, IVF and microinjection of thawed
fertilized oocytes with CRISPR-Cas reagents
|
Gene therapy protocols with CRISPR-Cas
technologies.![]()
These are some of the relevant articles
published reporting the use of CRISPR-Cas tools in gene therapy
procedures, in vitro (on cells) and in vivo (on animals), at PRE-CLINICAL
level. This is
one of the fastest growing applications of CRISPR-Cas approaches.
|
CRISPR-Cas
attempts in human embryos i.![]()
These are the first attempts of using
CRISPR-Cas technology in human embryos, logically encountering
similar results as reported in other mammalian species, that is:
mosaicism and potential off-targets. Nothing new, but important to
be aware of, in order to carefully think before considering the
serious use of CRISPR-Cas in human embryos, an approach currently
not recommended. Articles commenting these approaches are also added
to this list.
|
CRISPR
approaches in genome-wide screenings![]()
These are some of the relevant articles
published reporting the use of CRISPR-Cas tools in genome-wide
screening applications.
|
Which starting plasmids do I
need to obtain/purchase to begin using CRISPR-Cas9 technology?.![]()
There are currently many plasmids with
multiple Cas9 variants and diverse templates for preparing the
synthetic small guide RNA (sgRNA). We routinely use and recommend
start using the following plasmids, all available through Addgene.
All these plasmids do not contain the T7 promoter in their backbones. Hence, this
feature must be added at the 5' of the oligonucleotide used to
amplify the DNA region to be transcribed into RNA.
|
Other Cas9 proteins for
genome editing experiments.![]()
Cas9 was originally obtained from
Streptococcus pyogenes, but it exists in many other bacteria. In
addition, new variants, with novel properties, are being isolated
and characterized.
|
The new Cpf1
(Cas12a) nuclease and its derivatives.![]()
There are other CRISPR-Cas-like systems not using Cas9 as
the effector-nuclease protein. One of the first ones being
characterized was Cpf1, a single RNA-guided Endonuclease described
by Feng Zhang's lab in 2015. Here we will be listing relevant
publications describing the new properties of this new nuclease as they are released. Cpf1 has been known re-named to Cas12a.
|
Additional CRISPR-Cas-like systems not using Cas9 as
the effector-nuclease protein. C2c2 nuclease has been now re-named to Cas13a.
|
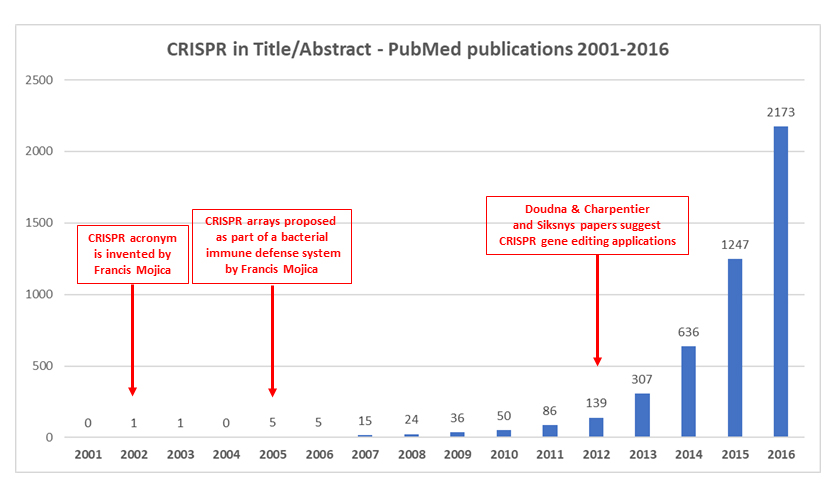
Graphic illustrating the evolution of CRISPR publications in PubMed (by Lluís Montoliu ©)
Other sources of Cas9 for
genome editing experiments.![]()
Cas9 is normally provided to cells as an
expression vector, or, to embryos, as purified in vitro-transcribed
RNA from any of the existing plasmids. However, there are additional
sources for Cas9 worth taking them into account, such us the use of
Cas9 mice (constitutively or conditionally expressing a Cas9-expressing
cassette), Cas9 lentiviral vectors or Cas9 purified protein itself.
|
Targeting
and Editing RNA with CRISPR.![]()
New CRISPR-related reagents have
been reported with new endonucleases with novel properties,
including RNA-dependent RNA endonucleases, where the CRISPR system
can be used for targeting and editing RNA, and not DNA. Some of the
first papers describing these new nucleases and their applications
are included in this non-exhaustive list.
|
How do I choose the short DNA
sequences from my favourite gene to design the sgRNA?.![]()
There are several CRISPR-Cas9 web sites
providing extremely useful services and helping you to design the
sgRNA complementary to the expected target site within your
favourite gene or genomic location. These are the ones we currently
use and recommend using:
Breaking-Cas is a most flexible new guide designing web tool, developed at CNB. This program is useful for predicting guide RNAs for SpCas9, SaCas9, AsCpf1, FnCpf1, C2c2 and other newer nucleases, like NgAgo, using all genomes (>1000) available from Ensembl, and allowing to place any given PAM sequence at 5' or 3', or no PAM at all (for Ago), any length of target sequence, any experimental data regarding frequencies of nucleotides per site, etc...
|
Where can I get full
protocols for using CRISPR-Cas-related technologies?.![]()
There are different publications and web
resources where you can find detailed protocols for using the CRISPR-Cas9
techniques. These are some of the ones we recommend using:
|
Successful
Knock-In strategies with CRISPR-Cas tools.![]()
There are different publications where
diverse strategies resulting in succcessful Knock-In mutant alleles
in animals have been reported.
|
CRISPR
experiments in livestock.![]()
CRISPR tools have boosted the generation of
gene-edited animals beyond the usual rodent laboratory models.
Numerous livestock species have been already successfully edited
using CRISPR approaches, thereby generating new and interesting
models for improving disease resistance, for adopting new productive
traits or for animal welfare considerations. Here are some of the
recent articles where CRISPR tools have been instrumental in the
creation of gene-edited livestock.
|
CRISPR and
gene drive approches.![]()
CRISPR tools have been also engineered to
trigger gene drive strategies, in which the inserted cassette
encodes both for the Cas9 nuclease and for the guide RNAs that
target the cassette to a given genomic locus, thus resulting in
biased non-mendelian inheritance. This approach has been mostly
explored for pest control in insects known to act as vectors for
infectious diseases, such as malaria, yellow fever and zika.
|
Anti-CRISPR and
Cas inhibitors.![]()
CRISPR-Cas tools are extremely efficient.
Anti-CRISPR proteins and Cas-inhbitors, as proteins or small
molecules, are beginning to be discovered and are expected to
provide a new source for controlling CRISPR activity. Some of the
recent developments are described here.
|
CRISPR-Cas tools can trigger off-target
mutations, depending on many parameters, most importantly the RNA
guide used. The existence and relevance of these potential off-target
altered genomic sequences is often reported and a matter of
discussion. Some of these papers are summarized here.
|
CRISPR experiments can be associated with
several Ethics issues, including its application to animals, to
human beings, to human embryos and to wild-type species in the
environment. A number of Ethics debates have been triggered over the
past years around these issues, some of these documents are listed
here:
|
The following list is a NON-exhaustive
compilation of Prizes awarded to several key researchers in this
field for their various contributions to our current knowledge on
the CRISPR systems and CRISPR tools for gene editing purposes
|
Scheme illustrating the most advanced CRISPR-Cas9 reagent: the RiboNucleoProtein (RNP) (by Lluís Montoliu ©)
Commercial
resources for CRISPR-Cas-related reagents.![]()
The following list is a NON-exhaustive
compilation of companies producing CRISPR-Cas-related reagents
commercially, fulfilling the need for intermediate reagents (i.e.
longer DNA oligonucleotides) of final reagents (i.e. RNAs) ready-to-use
for CRISPR-Cas applications. If you know additional companies
marketing similar products, please do let us know.
|
Where can I discuss with
other colleagues about the use of CRISPR-Cas9 techniques?.![]()
There are several lists, forum and google
groups where the use of CRISPR-Cas9 techniques is regularly
discussed. These are the ones we use:
|
Global
sites of CRISPR information, publications & commentaries.![]()
| There are several global sites of CRISPR informatiuon, publications and commentaries, including: |
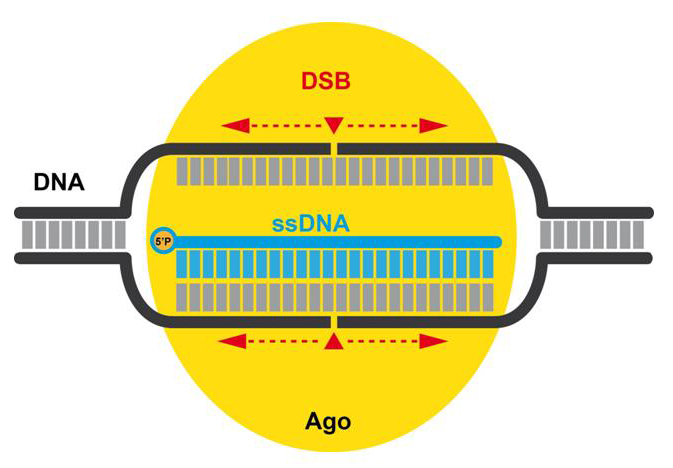
Beyond CRISPR-Cas:
Ago (Argonaute)-no evidences of gene editing activity .![]()
These are the first papers reporting a new
(what would have been possibly the fourth, after ZFN, TALEN and CRISPR)
promising gene editing tool: Ago (Argonaute), also from prokaryotes,
a DNA-guided DNA endonuclease. Current
evidences collected world wide do not confirm the results
initially reported in Nature Biotechnology by Gao et al. (2016) and
do not support any proof of gene editing activity that can be
attributed to Ago.
Using
Gaetan Burgio's last sentence in his latest preprint : "Together
our findings indicate that NgAgo is unsuitable as a genome editing
tool." Eventually, the original paper in Nature
Biotechnology has been retracted by the authors on 2 August 2017.
|
Scheme illustrating the pressumptive Ago mechanism of action (by Lluís Montoliu ©)